- Services
Services
Inoviem provides a full range of services from drug discovery to clinical development by leveraging its platforms and direct access to human pathological specimens.
All the platforms and services are label-free with no modification of the original molecules.Unravel disease relevant Mode of Action (MoA) of your molecule directly on human tissues.
Identify and validate targets in your study model or in Human.
Thanks to our ex-vivo pharmacology expertise we identify the best disease and patients subgroups.
Using patients’ samples and PIMS® technology, we identify those individuals who can benefit best from your compound as soon as the early stage of preclinical studies.
Compounds of interest can be profiled for their drugability based on their mode of action and efficacy.
Translational pharmacology empowers your ability to identify biomarkers and advance clinical developments.
Advanced SPR and proteomics drive precise analysis of biomolecular interactions and protein landscapes.
Meticulous collection and biobanking of human samples fuel robust, translational research.
- Platforms
Platforms
Inoviem provides breakthrough protein technologies for each phase of the drug development process.
One of the major advantages of our label-free technologies used under physiological conditions and in human tissue – NPOT® and PIMS® – is that the test compound retains its original structure, as well as the exact same molecular structure that would be used in therapyPIMS®
Designed to study molecular interactions and predict therapeutic responses in biomedical research
Biomarkers discovery
Methodology for the proteomic profiling of human individuals in the context of translational evaluation of biomarkers and clinical development of new drug candidates.
- Knowledge centre
- About us
About us
Inoviem Scientific is a Bioanalytical R&D company established in 2011 in Strasburg – France.

Our mission
Our mission is clear: to get the right treatment to the right patients within the right therapeutic window.
- Contact us
Background
We applied for the first time two label-free technologies, physiological intermolecular modulation spectroscopy (PIMS®) and nematic protein organization technique (NPOT®) in anti-TNF refractory IBD patients to identify clinical responders to vedolizumab therapy and elucidate their underlying functional molecular network.
Methods
PIMS® analysis was performed in peripheral blood taken prior to the first vedolizumab application in 20 IBD patients (Crohn’s disease n=13; ulcerative colitis n=7) refractory to at least one previous anti-TNF agent therapy. Peripheral blood taken from clinical responders and non-responders at week 14 of vedolizumab therapy were additionally subjected to NPOT® analysis. Response to therapy was assessed by respective clinical disease activity scores (partial Mayo Score and Harvey-Bradshaw-Index).
Results
Clinical response to vedolizumab treatment was observed in 7 of 13 Crohn’s disease and 4 of 7 ulcerative colitis patients at week 14. Response to therapy was accurately predicted by PIMS® blood analysis in 100 % of ulcerative colitis and 77% of Crohn’s disease patients. Overall prediction of clinical response with PIMS® blood analysis was achieved with a 89% positive predictive value (PPV) and a 82% negative predictive value (NPV). NPOT® analysis revealed the heightened expression of the proteins ITGB7, ITGAV, ITG3, PF-4 and ASGH in the peripheral blood of vedolizumab responders compared to non-responders.
Conclusion
PIMS® analysis of the blood of anti-TNF refractory IBD patients was able to stratify responders to vedolizumab therapy with high accuracy and specificity. NPOT® technology could decipher underlying molecular networks in the blood of responders, enabling subsequent personalized therapeutic approaches in IBD.
Of the 13 vedolizumab-treated Crohn’s disease patients, 7 were considered as responders and 6 as non-responders at week 14 of therapy (Table 1).
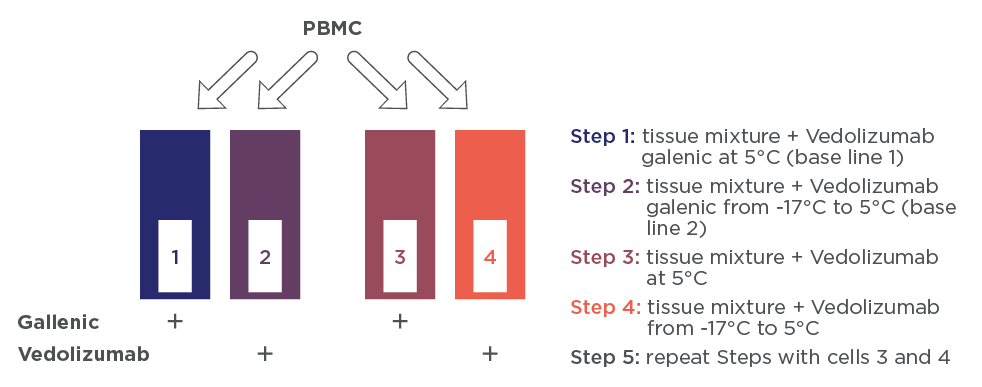
Figure 1 PIMS® experimental steps. All experiments are performed using the double beam PIMS® instrument. The boxes represent the PIMS cells. All cells contain the mixture of PBMCs. Cell 1 and 3 contain only the excipients (L-histidine chlorhydrate, L-histidine monohydrate, L-arginine chlorhydrate 18, saccharose polysorbate 80) of vedolizumab, whereas cells 2 and 4 contain vedolizumab in its galenic formulation.
Of the 7 vedolizumab-treated ulcerative colitis patients, 4 were considered as responders and 3 as non-responders at week 14 of therapy (Table 2).
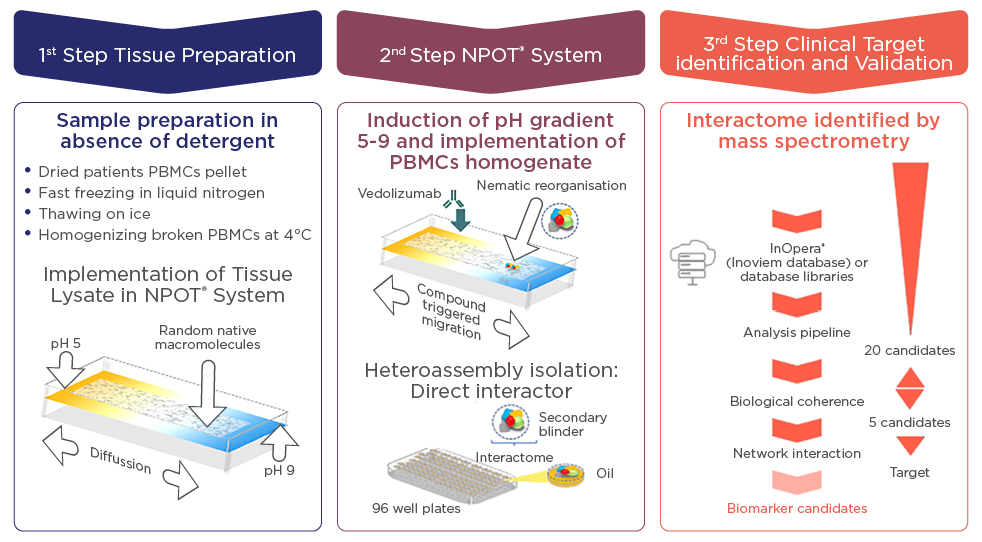
Figure 2 Schematizing NPOT® experiment using patients PBMCs and vedolizumab. The first step describe the sample preparation until loading into the NPOT® with a pH gradient; the second step illustrates the specific heteroassemblies formation and isolation. The resulting heteroassemblies protein contents are analyzed by liquid chromatography coupled with tandem mass spectrometry (LC−MS/MS).
Profile of the Individual Macromolecular Volume (IMV) during PIMS® of an IBD patient non-responder (A and B) and responder (C and D) to vedolizumab treatment are depicted in Figure 3.
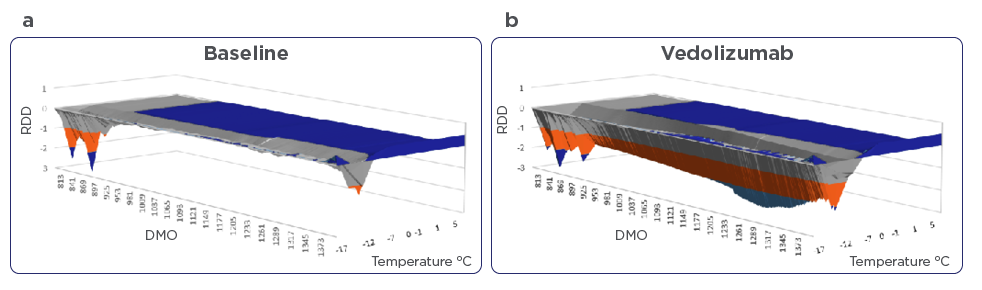
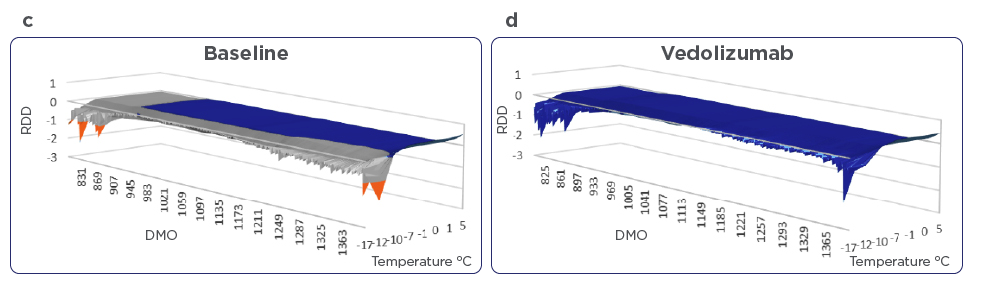
Figure 3: Profile of Individual Macromolecular Volume (IMV) of an IBD patient non-responder (A and B) and responder (C and D) to vedolizumab therapy. In non-responder patients, there was no modulation from the baseline of patients PBMCs macromolecular resonance in presence of vedolizumab. In responder patients, PBMCs macromolecular resonance was modified in the presence of vedolizumab. Note the change in the molecular oscillation in presence of vedolizumab as the temperature rises from -17°C to 5°C. For calculating the DMO and RDD, the entire results from -17 to 5°C are taken into account. DMO= differential molecular oscillation, RDD= relative dynamic diffraction.
These profiles are generated from the continuous sampling of define Near Infrared spectra of patient’s PBMC homogenates as the temperature rises from -17°C to 5°C. The obtained IMV is the result of the difference between the signal from the test cell and the blank. A negative IMV means that the blank cell has higher molecular resonance compared to the test cell. In non-responder patients, there was no change in IMV in presence of vedolizumab. This shows the lack of the antibody’s impact on molecular resonance whereas in responder patients, vedolizumab did impact the molecular resonance and therefore altered the IMV.
Prior to deblinding of data, PIMS®-treatment predictions were documented for each patient. PIMS® analysis of blood taken at week 0 was in accordance with clinical response to vedolizumab treatment at week 14 in 77% of vedolizumab treated Crohn’s disease patients (n=13). In vedolizumab treated ulcerative colitis patients (n=7), we found PIMS® analysis in accordance with clinical response at week 14 in 100% of patients. The correlation between PIMS® prediction and clinical response data were calculated as Positive Predictive Value (PPV) and Negative Predictive Value (NPV) for all IBD patients (n=20) which were 89 % and 82 %, respectively.
For NPOT® experiments, three IBD patients from the responder and non-responder group were randomly selected. In the group of 3 responders, there were 1 Crohn’s disease and 2 ulcerative colitis patients. In the group of non-responders, there were 2 Crohn’s disease and 1 ulcerative colitis patients. Blood taken at week 14 of these six patients were subjected to NPOT® analysis. The obtained heteroassamblies from responders and non-responders are shown (Figures 4A and 4B).
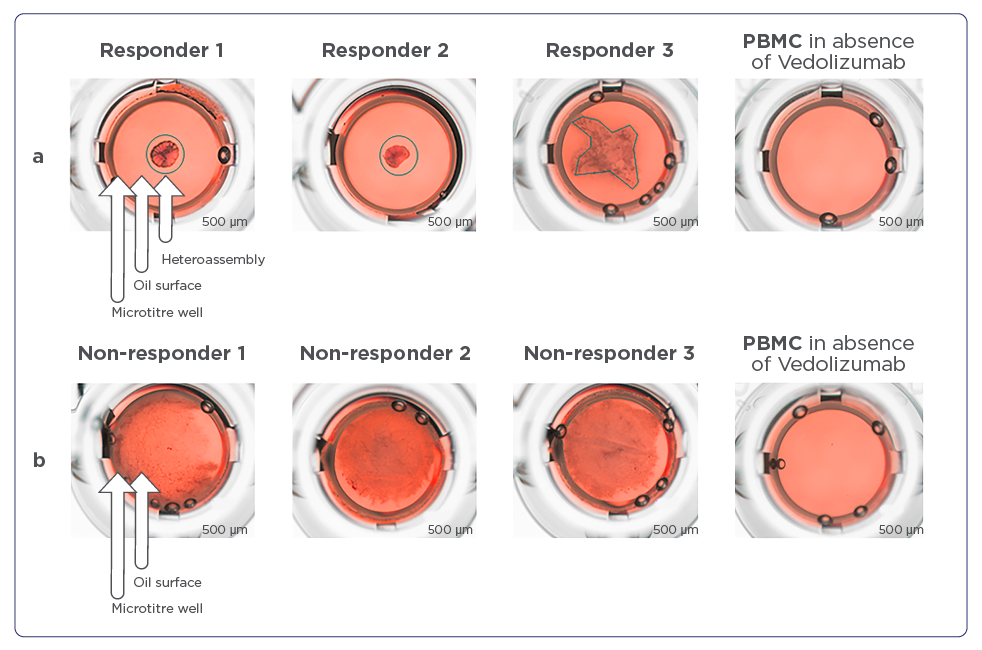
Figure 4 A and B:
A) Heteroassemblies of three vedolizumab responders as assessed by NPOT® analysis. Heteroassemblies are depicted by black circles or black lines. Each patient’s PBMC extract was put in contact with vedolizumab and subsequently gave rise to clearly-defined heteroassemblies with common reticular morphology and two out of three patients with symmetric and one with asymmetric forms. There are no heteroassemblies formed in the absence of vedolizumab.
B) Heteroassemblies of three non-responder patients to vedolizumab. Heteroassemblies are diffused with no distinguishable shape. There are no heteroassemblies formed in the absence of vedolizumab.
Heteroassamblies from responder patient’s PBMCs homogenates at week 14 were well structured and had distinct forms. Proteomic analysis using LC/MS-MS sequencing identified n=523, n=635, and n=598 proteins respectively from each analysed IBD patient’s PBMC homogenates. The analysis after filtering the frequent hits as well as non-specific binding, revealed 23 proteins found to be specific for vedolizumab treated clinical responders. These proteins were subjected to the string database analysis to form the interactome.
Of 23 proteins, 13 build a distinct network. The network consists of the proteins ITGAV (integrin alpha V), ITGB3 (integrin beta 3), AHSG (alpha2-HSglycoprotein), PF4 (platelet factor 4), PPBP (platelet basic protein), GP9 (platelet glycoprotein IX) and GC (vitamin D-binding protein). Of these 7 proteins, ITGAV, ITGB3 and ASHG interact directly with the target ITGB7 (alpha4beta7) whereas the other four proteins PF4, PPBP, GP9 and GC interact indirectly with ITGB7 (Figure 5).
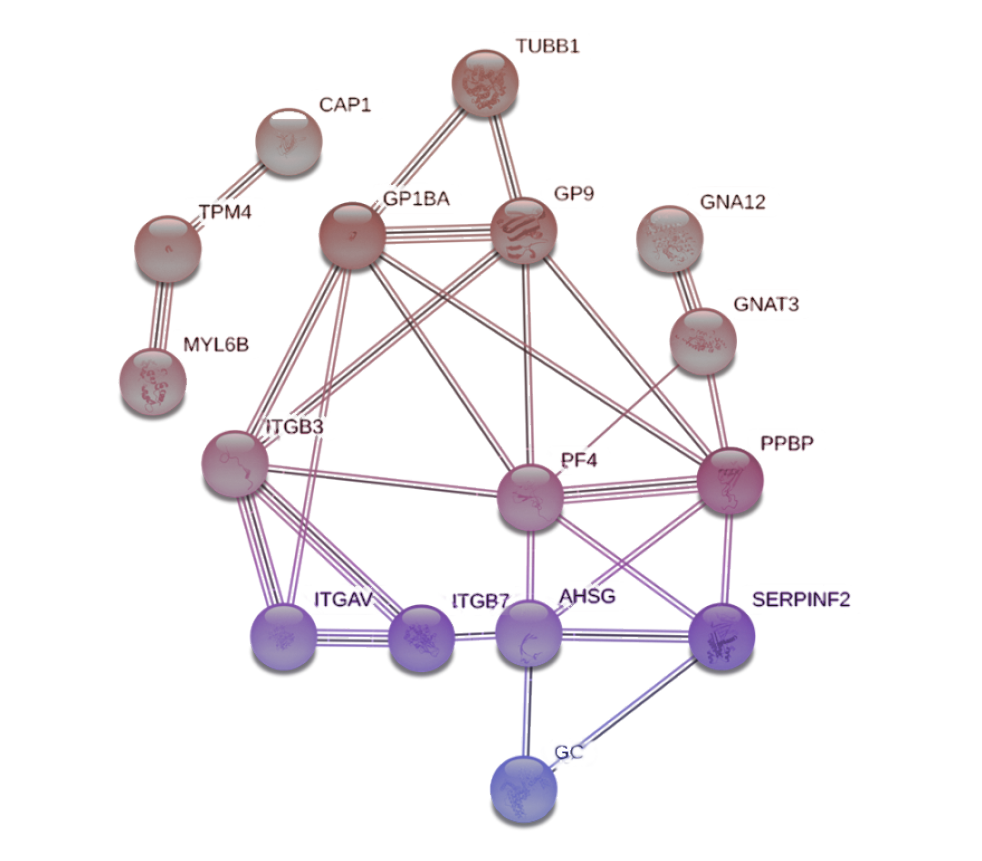
Figure 5: STRING network from responder patients to vedolizumab.
NPOT® heteroassemblies protein content was analyzed using STRING. The near interactor of signaling pathway is comprised of ITGB7, ITGAV, ITGB3, AHSG and PF4 (encircled proteins)
Heteroassamblies from non-responder patient PBMCs homogenate at week 14, contrary to responders, were diffuse and floating on the oil surface (Figure 6). Proteomic analysis using LC/MS-MS sequencing identified n=839, n=589 and n=569 proteins respectively from each IBD patient’s PBMC homogenate. After filtering the frequent hits as well as non-specific binding, 10 proteins were found to be specific for vedolizumab, which also included its target ITGB7. In comparison to responder patients, no-distinct interaction network could be established (Figure 6).
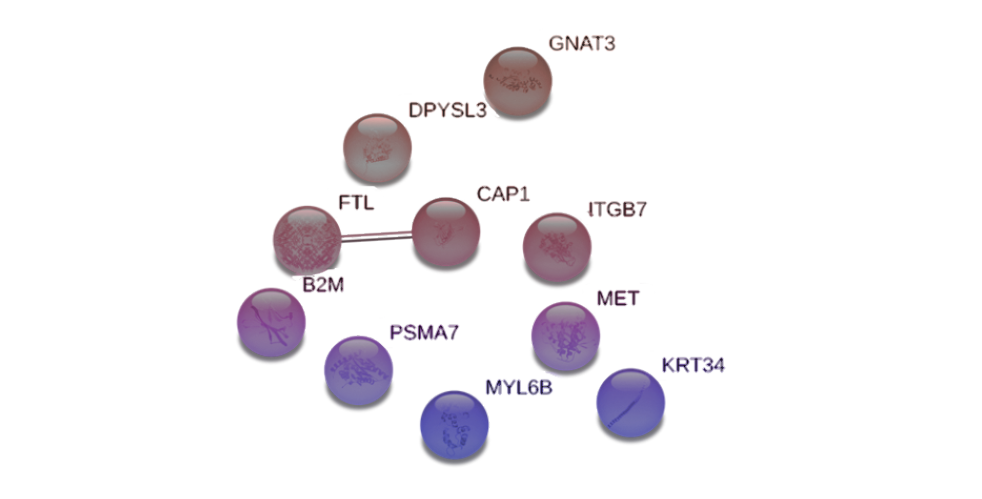
Figure 6: STRING network from non-responder patients to vedolizumab.
There is an absence of interactors and therefore a signaling pathway could not be identified.
DISCUSSION
Taken together, this prospective study demonstrates the capacity of label-free based proteomic approaches to predict clinical efficacy of vedolizumab therapy in an individual manner and pave the way for identification of its success predictors. Based on the reported observations and subsequent validation, PIMS® and NPOT® analysis might be applied in disease management of anti-TNF non-responders, to decide if subsequent biological treatment with vedolizumab should be initiated or if another therapeutic option (e.g. ustekinumab) should be favored instead. In parallel to the preparation of the follow-up study, the clinical values of GC and PF4 as successful biomarkers for vedolizumab therapy will be assessed in the blood of the treated patients. This could open the opportunity to develop a companion test for prediction of vedolizumab efficacy in IBD patients and enable a much needed personalized therapeutic approach, especially in IBD patients failing first line therapy with an anti-TNF antibody.
References
- Kaplan GG. The global burden of IBD: from 2015 to 2025. Nat Rev Gastroenterol Hepatol. 2015;12:720–7.
- Abraham C. and JH Cho. Inflammatory Bowel Disease. N Engl J Med. 2009; 361: 2066-2078.13.
- Eftekhari P, Glaubitz L, Breidert M, et al. Physiological intermolecular modification spectroscopy for the prediction of response to anti-tumor necrosis factor therapy in patients with inflammatory bowel diseases. Dig Dis. 2014; 32:446–54.20. Cheung MS, Klimov D, Thirumalai D. Molecular crowding enhances native state stability and refolding rates of globular proteins. PNAS. 2005; 102:4753–8.
- Brovchenko I, Geiger A, Oleinikova A. Liquid-liquid phase transitions in supercooled water studied by computer simulations of various water models. J Chem Phys. 2005; 123: 044515.
- Brovchenko I, Geiger A, Oleinikova A, et al. Phase coexistence and dynamic properties of water in nanopores. Eur Phys J E Soft Matter. 2003;12:69–76.
- Tanford C. The Location of Electrostatic Charges in Kirkwood’s Model of Organic Ions. J Am Chem Soc 1957; 79:5348–52.
- Richter K, Nessling M, Lichter P. Macromolecular crowding and its potential impact on nuclear function. Biochim Biophys Acta Mol Cell Res. 2008; 1783:2100–7.
- Dobson CM. Protein folding and misfolding. Nature 2003;426:884–90.
- Beyrath J, Pellegrini M, Renkema H ,et al. KH176 safeguards mitochondrial diseased cells from redox stress-induced cell death by interacting with the thioredoxin system/peroxiredoxin enzyme machinery. Sci Rep. 2018; 8:6577.
- Walf-Vorderwülbecke V, Pearce K, et al. Targeting acute myeloid leukemia by drug-induced c-MYB degradation. Leukemia. 2018; 32:882–89.
- Wang L, Eftekhari P, Schachner D, et al. Novel interactomics approach identifies ABCA1 as direct target of evodiamine, which increases macrophage cholesterol efflux. Sci Rep. 2018; 8:11061.
